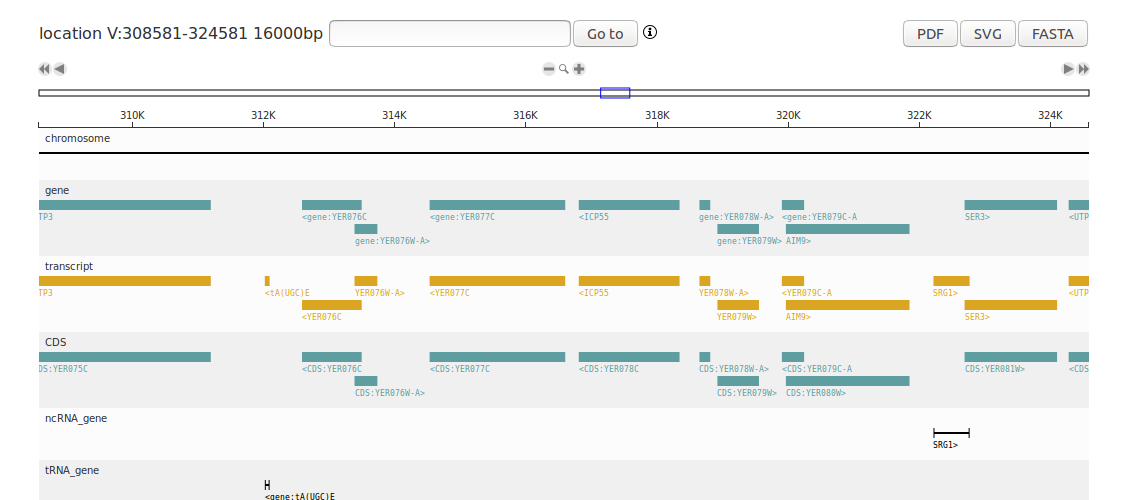
This tutorial shows how to create a D3GB genome browser from GFF and Fasta files with the R package. This will create a complete genome browser of Saccharomyces cerevisiae. Install the D3GB package and write the following code in R:
library(D3GB)
# Download fasta file
fasta <- tempfile()
download.file('http://ftp.ensembl.org/pub/release-84/fasta/saccharomyces_cerevisiae/dna/Saccharomyces_cerevisiae.R64-1-1.dna.toplevel.fa.gz',fasta)
# Genome browser generation.
gb <- genomebrowser(getAssemblyFromFasta(fasta))
genome_addSequence(gb,fasta)
# Download gff file and add to the genome browser
gff <- tempfile()
download.file('http://ftp.ensembl.org/pub/release-84/gff3/saccharomyces_cerevisiae/Saccharomyces_cerevisiae.R64-1-1.84.gff3.gz',gff)
genome_addGFF(gb,gff)
# It creates a genome browser ready to be viewed in your browser.
plot(gb)
view result