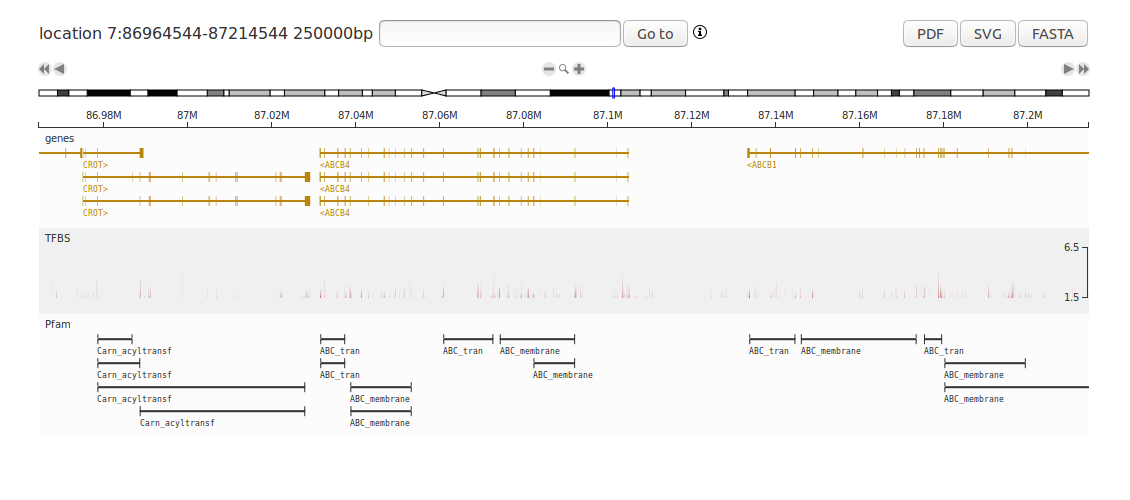
This tutorial shows how to create a complete human genome browser with the D3GB Python module. This example creates the genome browser and adds several tracks such as RNASeq expression, GWAS results, variants from exome sequencing samples, Variant Effect Predictor (VEP) annotations of variants, protein domains and gene annotations. Install the D3GB module and write the following code in Python 3:
import D3GB, urllib.request
# Genome browser generation.
# It creates a genome browser ready to view in a Firefox browser.
# For a server version, ready to be shared with Apache as a Website, set server=True
gwas, headers = urllib.request.urlretrieve('http://d3gb.usal.es/docs/data/GWAS_Catalog')
gb = D3GB.genomebrowser('GRCh37.bands', mapTrack = gwas, directory = 'HumanGenomeBrowser')
# Genes track
genes, headers = urllib.request.urlretrieve('http://d3gb.usal.es/docs/data/refFlat')
gb.addTrack(genes, 'refFlat', 'exons', '#B8860B')
# PFAM track
pfam, headers = urllib.request.urlretrieve('http://d3gb.usal.es/docs/data/Pfam')
gb.addTrack(pfam, 'Pfam', 'domain', '#303030')
# GWAS track
gb.addTrack(gwas, 'GWAS_Catalog', 'value', '#A52A2A', [0,100])
# Adding vep annotated vcf
vcf, headers = urllib.request.urlretrieve('http://d3gb.usal.es/docs/data/trio_vep')
gb.addVCF(vcf,'trio_vep',('Existing_variation','Consequence','IMPACT','GMAF'))
# Adding tissue expression
for i in ('adipose_tissue', 'adrenal_gland', 'brain', 'heart', 'kidney', 'liver', 'lung', 'ovary', 'pancreas', 'sigmoid_colon', 'small_intestine', 'spleen', 'testis'):
tissue, headers = urllib.request.urlretrieve('http://d3gb.usal.es/docs/data/'+i)
gb.addTrack(tissue,i,'score')
# Genomic sequence loading
fasta, headers = urllib.request.urlretrieve('ftp://ftp.ensembl.org/pub/grch37/release-85/fasta/homo_sapiens/dna/Homo_sapiens.GRCh37.dna.primary_assembly.fa.gz')
gb.addSequence(fasta)
view result